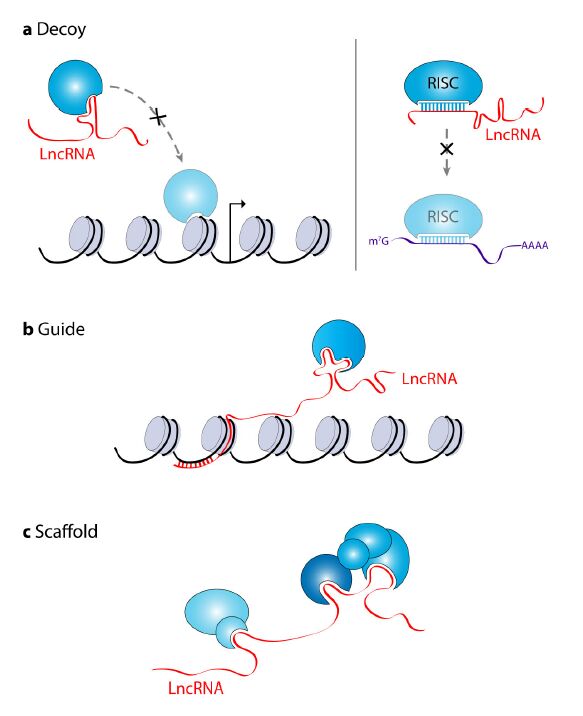
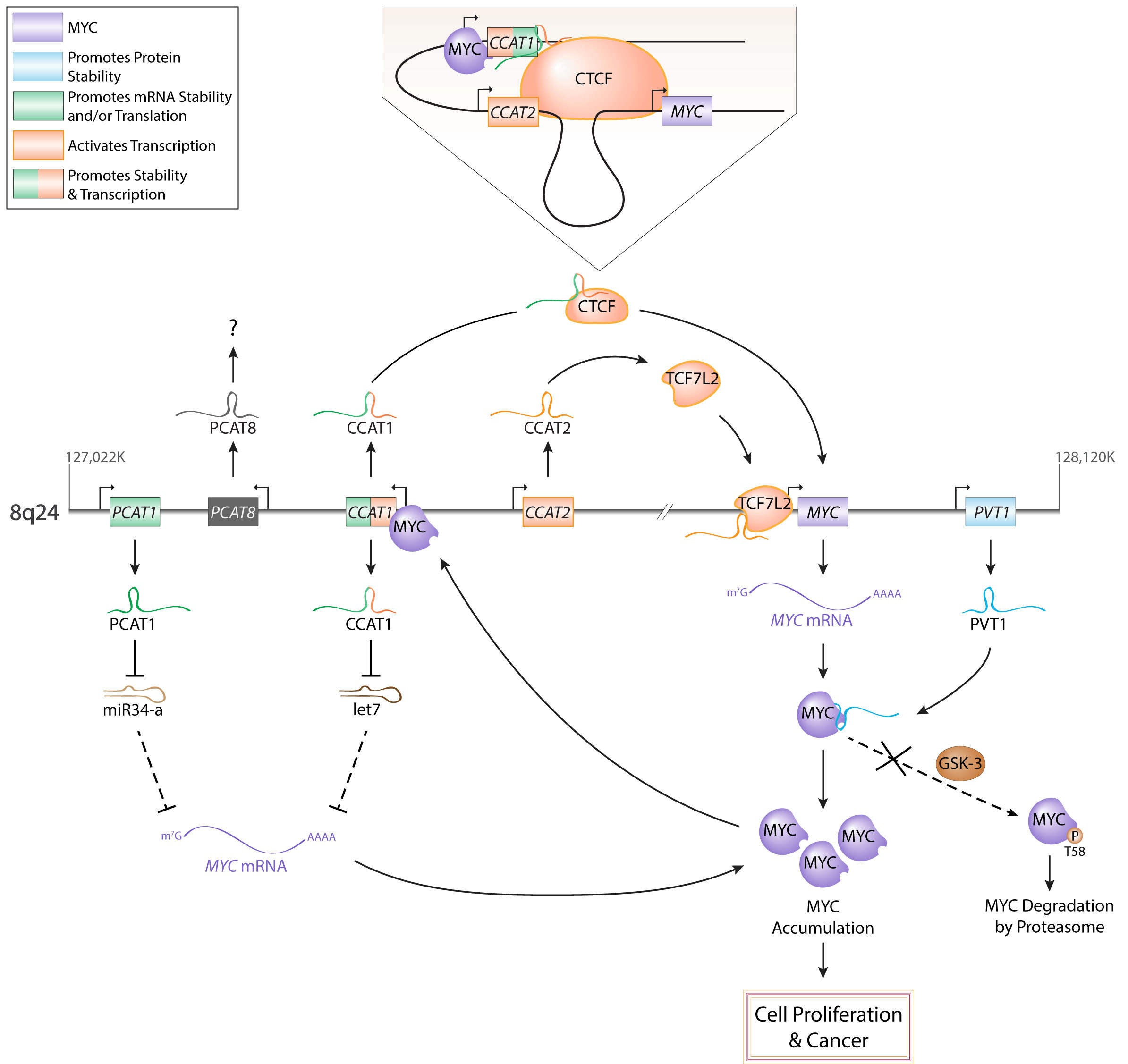
Long non-coding RNAs (lncRNAs) are a class of RNA molecules that are changing how researchers view eukaryotic gene regulation. Once considered to be non-functional products of low-level aberrant transcription from non-coding regions of the genome, lncRNAs are now viewed as important epigenetic regulators and several lncRNAs have now been demonstrated to be critical players in the development and/or maintenance of cancer. Similarly, the emerging variety of interactions between lncRNAs and MYC, a well-known oncogenic transcription factor linked to most types of cancer, have caught the attention of many biomedical researchers. Investigations exploring the dynamic interactions between lncRNAs and MYC, referred to as the lncRNA-MYC network, have proven to be especially complex. Genome-wide studies have shown that MYC transcriptionally regulates many lncRNA genes. Conversely, recent reports identified lncRNAs that regulate MYC expression both at the transcriptional and post-transcriptional levels. These findings are of particular interest because they suggest roles of lncRNAs as regulators of MYC oncogenic functions and the possibility that targeting lncRNAs could represent a novel avenue to cancer treatment. Here, we briefly review the current understanding of how lncRNAs regulate chromatin structure and gene transcription, and then focus on the new developments in the emerging field exploring the lncRNA-MYC network in cancer.
1.
Introduction
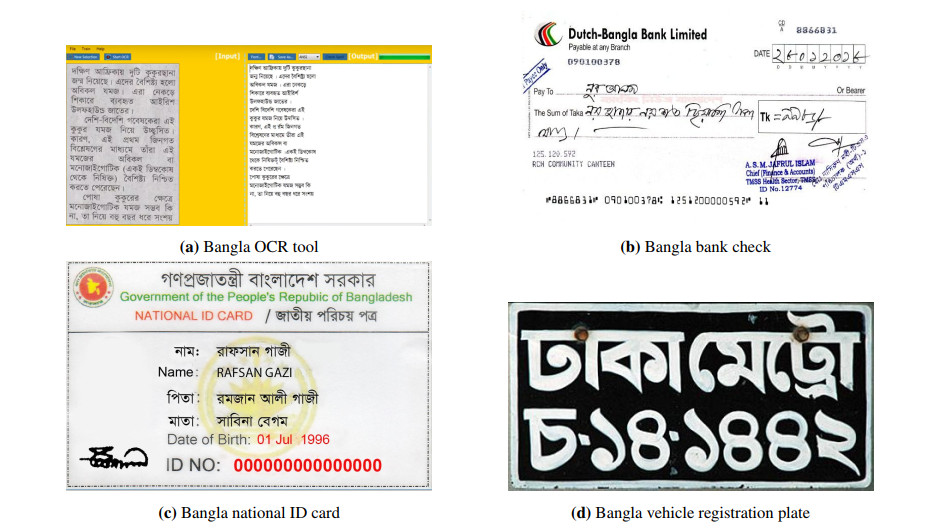
Printed script letter classification has a tremendous commercial and pedagogical importance for book publishers, online Optical Character Recognition (OCR) tools, bank officers, postal officers, video makers, and so on [1,2,3]. Postal mail sorting according to zip code, the verification of signatures, and check processing are usually done with the application of grapheme classification [4,5,6]. Some sample images of its application are shown in Figure 1.
Statistically, the importance of printed script letter classification is enormous when a large population uses a specific language. For example, with nearly 230 million native speakers, Bengali (also called Bangla) ranks as the fifth most spoken language in the world [7]. It is the official language of Bangladesh and the second most spoken language in India [8] after Hindi.
Handwritten character classification or recognition is particularly challenging for the Bengali language, as it has 49 letters and 18 potential diacritics (or accents). Moreover, this language supports complex letter structures created from its basic letters and diacritics. In total, the Bangla letter variations are estimated to be around 13, 000, which is 52 times more than the English letter variations [9]. Although several language grapheme classifications have received much attention [10,11,12], Bangla is still a relatively unexplored field, with most works done in the detection of vowels and consonants. The limited progress in exploring the Bengali language has motivated us to classify the Bengali handwritten letters into three constituent elements: root, vowel, and constant.
Previously, several machine learning models have been used for different language grapheme recognition [13]. Several research using the deep convolutional neural network has been successful in the detection of handwritten characters in Latin, Chinese, and English language [14,15]. In other words, successful feature extraction became possible using different types of layers in neural networks. The convolutional neural network with the augmentation of the images of handwriting can generate a better model through training in deep learning [16,17]. Previous research in this area faced fewer challenges regarding variations, massive data usage, and model creation that deep learning needs most regarding the high number of label classification [19,20].
This paper proposes a deep convolutional neural network with an encoder-decoder to facilitate the accurate classification of Bangla handwritten letters from images. We use 200, 840 handwritten images to train and test the proposed deep learning model. We train in 4 steps with four subsets of images containing 50, 210 images each. In doing so, we create three different labels (roots, vowels, and consonants) from each handwritten image. The result shows that the proposed model can classify handwritten Bangla letters as follows: roots-93%, vowels-96%, and consonants-97%, which is much better than previous works done on Bangla grapheme variations and dataset size.
The paper is organized as follows: Section 2 discusses the brief background of existing research on this research area. Section 3 is devoted to present Bangla handwritten letter dataset details. This section discusses the dataset structure and format and augments the dataset to create many variations. Section 4 discusses the architecture of the models for Bangla handwritten grapheme classification, and in section 5, we discuss the experimental process, tools, and used programming language. Section 6 shows the detailed results of the training process and validation. Finally, section 7 concludes the research work by discussing the contributions and applicability in the classification of Bangla handwritten letters.
2.
Related work
Optical Character Recognition is one of the favorite research topics in computer graphics and linguistic research. This section briefly discusses two areas first, how deep learning is used in character recognition, second, how deep learning is used in Bangla handwritten digit and letter classification.
In optical character recognition, many research works have been proposed. Théodore et al. showed that learning features with CNN is much better for handwritten word recognition [21]. However, the model needs a longer processing time when classifying a word compared to a letter. Zhuravlev et al. mentioned this issue in their research and experimented with a differential classification technique on grapheme images using multiple neural networks [22,23]. However, the model works better with that small dataset and will fail to detect when augmented images will be provided. Jiayue et al. discussed this issue and proved that proper augmentation before feeding into CNN could be more efficient in grapheme classification [24].
Regarding Bangla character and digit recognition, there are few kinds of research available. A study by Shopon et al. presented unsupervised learning using an auto-encoder with deep ConvNet to recognize Bangla digits [25]. A similar study by Akhand et al. proposed a simple CNN for Bangla handwritten numeral recognition [26,27]. These methods achieved 99% accuracy in detecting Bangla digits and faced fewer challenges in classifying only ten labels than more character recognition labels.
A study on Bangla character recognition by Rabby et al. used CMATERdb and BanglaLekha as datasets and CNN to recognize handwritten characters. The resulted accuracy of CNN was 98%, and 95.71% for each dataset [28]. Another study by Alif et al. showed ResNet-18 architecture is giving similar accuracy on CMATERdb3, which is a relatively large dataset than previous [29,30]. The limitation of these two research is their image variations and dataset size.
This paper addresses the limitations of previous research in image augmentation, dataset size, proper model creation, and a high number of label classification.
3.
Dataset preparation
This section describes raw data and data augmentation to ensure better data preparation for our proposed model.
3.1. Data background
This study uses the dataset from the Bengali.AI community that works as an open-source dataset for research. The dataset was prepared from handwritten Bangla letters by a group of participants. The images of these letters are provided in parquet format. Each image is $ 137 \times 236 $ pixels. Some sample handwritten images are shown in Figure 2.
The Bengali language has 49 letters with 11 vowels, 38 consonants, and 18 potential diacritics. The handwritten Bengali characters consist of three components: grapheme-root, vowel-diacritic, and consonant-diacritic. To simplify the data for ML, we organize 168 grapheme-root, 10 vowel-diacritic, and 6 consonant-diacritic as unique labels. All the labels are then introduced as a unique integer. Table 1 summarizes the metadata information of the training dataset.
The raw training dataset has a total of 200, 840 observations with almost 10, 000 possible handwritten image variations. The raw testing dataset is created separately to distinguish it from the training dataset. Table 2 summarizes the metadata information of the test data.
3.2. Dataset augmentation
The raw dataset is a set of images in the parquet format, as discussed in the previous section. The images are generally created from a possible set of grapheme writing, but it does not cover all the aspects of writing variations. To create more variations (more than 10, 000), dataset augmentation becomes a necessary step. In reality, it has around 13, 000 possible letter variations that make the problem harder than any other language grapheme classification. Therefore, a pre-processing of the dataset is done to increase the more number of grapheme variations.
We apply the following data augmentation techniques: (1) shifting, (2) rotating, (3) changing brightness, and (4) applying zoom. In all cases, some precautions are taken so that augmented handwritten images are well generated. For example, too much shifting or too much brightness can lose the image pixels [31]. Applying random values to those operations is also prohibited during our pre-processing of the dataset.
In terms of shifting, we apply the following image augmentation: width shift, and height shift on our images. In rotation, an image was rotated to (1) 8 degree, (2) 16 degree, and (3) 24 degree in both positive and negative direction. In case of zoom, we apply (1) 0.15%, and (2) 0.3% zoom in. Some sample output of Bangla handwritten letter's augmentation is shown in Figure 3.
The augmenting was minimized to only four options due to minimizing the risk of false image creation. As our dataset is related to characters, we need to verify that augmentation may add false image augmentation. For example, there is a horizontal flip option for image augmentation. If we apply that to our dataset, some non-Bangla handwritten images and the proposed model may learn wrongly to classify the handwritten letters.
4.
Methodology
This section describes the architecture of the models that we build for Bangla grapheme image classification. We also discuss the neural networks and their necessary layers useful for fitting data into the model.
4.1. Neural network
A neural network is a series of algorithms that process the data through several layers that mimic how the human brain operates. A neural network for any input $ x $ can be expressed as:
where, $ w_j $ is the network weights, $ b $ is a bias term, and $ f $ is a specified activation function. For a better approximation, multiple neurons are used in the form of hidden layers as follows:
To obtain the final output with higher accuracy, multiple hidden layers are used. The final output can be obtained as:
For image processing, we flat the image and feed it through neural networks. A vital characteristic of the images is that images have high dimensional vectors and take many parameters to characterize the network. To solve this issue, convolutional neural networks were used to reduce the number of parameters and adapt the network architecture specifically to vision tasks. CNN's are usually composed of a set of layers that can be grouped by their functionalities.
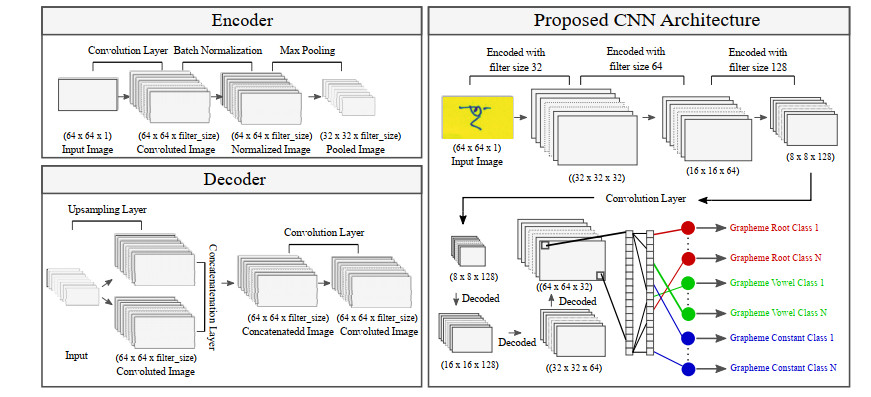
In this study, we implemented the CNN architecture with an encoder-decoder system. The encoder-decoder classifier works at the pixel level to extract the features. A recent work by Jong et al. shows that how encode-decoder networks using convolutional neural networks work as an optimizer for complex image data sets [33]. The encoder and decoder are developed using convolution, activation, batch normalization, and pooling layers. The detailed picture of these layers is described in the following section and shown in Figure 4.
4.2. Convolution layers
The convolution layer extracts feature maps from the input or previous layer. Each output layer is connected to some nodes from the previous layer [34,35] and valuable in the classification process. The convolutional layer is used to the sliding window through the image and convolves to create several sub-image, increasing the volume in terms of depth. The implementation view of the convolutional layer exists in both encoder and decoder, shown in Figure 4.
4.3. Activation layers
This section describes the activation function used in neural networks for proposed model training. We include the rectified Linear Unit (ReLU) function to add the non-linearity in the proposed network. The function is defined as
where it returns 0 for negative input and $ x $ for positive input.
There is some other nonlinearity function named as $ sigmoid $ and $ tanh $. The $ sigmoid $ non-linearity function can also be declared as follows
which maps a real number x to a value between [0, 1]. Another nonlinear function $ tanh $ maps the values into the interval [$ - $1, 1].
As the Bangla grapheme list contains more labels to detect than other language graphemes, we implement several non-linear layers as an encoder and a corresponding set of decoders to improve the recognition performance. However, we use ReLu due to its linear form, and this function improves the performance of classification compared to $ sigmoid $, and $ tanh $ functions [36,37]. An essential feature of such ReLu function is that it shows non-linearity around 0 because its slope is not constant around 0. Therefore, when the CNN model includes bias terms, the nodes can change the slope at different values for our input using the ReLu function [37].
In our model, we introduce such ReLu functionality after each non-pooling layer in the encoder to map the value of each neuron to an actual number. However, in some cases, such function can die during training. To solve this issue, leaky Relu has been added so that if there is any negative value, it will add a slight negative slope [38].
4.4. Normalization & dropout layers
The batch normalization layer normalizes each input channel across a mini-batch [39,40]. Our study adjusts and scales the previously filtered sub-images and normalizes the output by subtracting the batch mean and diving by the standard deviation of the batch. Then, it shifts the image input by a learnable offset. Generally, using such a layer after a convolution layer and before non-linear layers is useful in speeding up the training and reducing sensitivity to network initialization [39].
We also implement a dropout layer during the final classification step. We use this layer to reduce the labels by setting zero, which has less probability in classification [41,42].
4.5. Pooling layers
The pooling layer is mainly used to reduce the resolution of the feature maps. To be specific, this layer downsamples the volume along the image dimensions to reduce the size of representation and number of parameters to prevent overfitting. Our model uses a max-pooling layer in each encoder block, which downsamples the volume by taking max from each block.
5.
Experiment
The proposed model is trained with a configuration of the epoch, loss function, and batch size in the experiment. Also, the model contains trainable and non-trainable parameters. The trainable parameter size for the proposed model is 136, 048, 154, whereas the non-trainable parameter size is 448. There are thirteen convolution layers, three pooling layers, three normalization layers, and four dropout layers are used in our model. The experiment is implemented using the Python programming language with Keras [43], and Theano [44] library.
In terms of configuration, the proposed model uses 25 epochs with a batch training size of 128. To calculate the loss function, we use categorical cross-entropy for root, vowel, and constant classification. Mean Squared Error (MSE) is another metric to calculate loss function but categorical cross-entropy performance is better in classification tasks [45].
After developing the model in Python, we run it on an Intel (R) Core (TM) i7-7500U CPU @ 2.70 GHz machine with 16GB RAM. For both validation and training, the same batch size and epochs were used. The experimental results performance is calculated using accuracy and model performance evaluation metric. We also use a loss function to evaluate how well our deep learning model trains the given data. Both of these metrics are popular in classification tasks using deep learning [46,47,48].
6.
Results & discussion
In this section, we present the outcome of the model in terms of evaluation metrics. The proposed CNN method is applied in four different phases on four subsets of the grapheme dataset and produces the results. This way, we test how more Bangla handwritten letter images are helpful to produce better deep neural network models. However, conducting this research with more subsets of images will have computational and complexity challenges. In evaluation, the accuracy and loss of each epoch of training and validation are used.
6.1. Phase 1 results
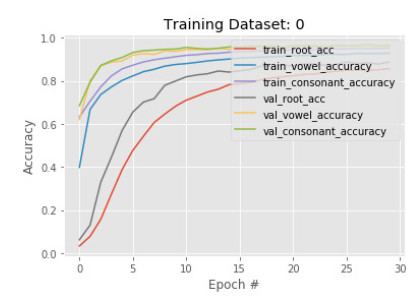
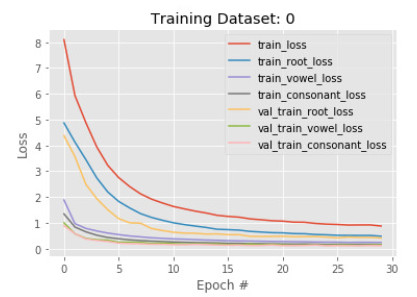
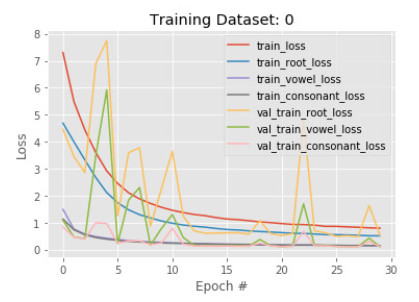
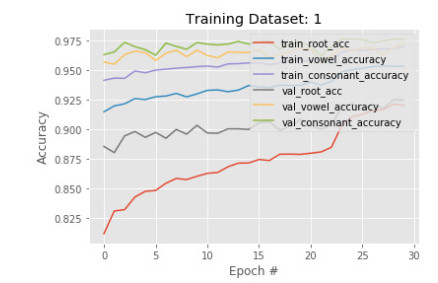
In the first phase, a subset of 50, 210 images is sent to training with 30 epoch. The results show that accuracy in detecting the root is less than the vowel and constant in both training and validation. The training and validation root accuracy are 85% and 88% respectively, where vowel accuracy is 92% and 95%, constant accuracy is 95% and 96%. This is because many root characters are needed to identify than the number of vowels, and constants are needed to identify. Figures 5 and 6 show the train and validation accuracy and cross-entropy loss over epochs. The results show that the model seems to have good converged behavior. It implies the model is well configured and no sign of over or underfitting.
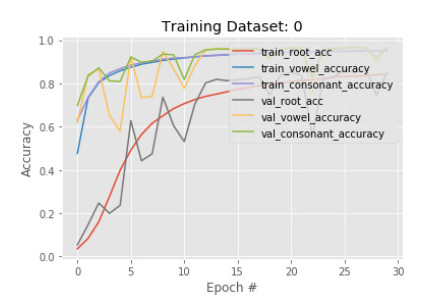
We also test how a CNN model with an encoder-decoder is compared to a traditional CNN that does not have an encoder and decoder concept. There are six convolution layers, three pooling layers, five normalization layers, and four dropout layers are used in the traditional CNN model. Figures 7 and 8 visualize the accuracy and loss of the simple CNN model for 30 epochs. The results we see from the figure that the loss and accuracy are fluctuating. The reason behind this, the simple CNN model is sensitive to noise and produces random classification results. This problem is also known as overfitting. The results show a better performance with an encoder and decoder concept than a traditional one.
6.2. Phase 2 results
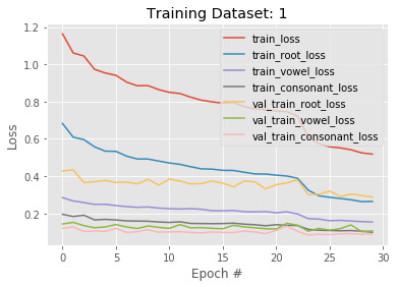
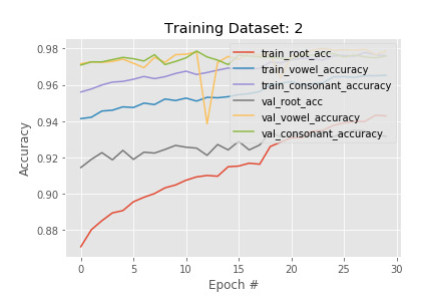
After getting a training accuracy of 85% in the first phase, we train another subset of images with the existing model. The hypothesis is that the more variations of images are trained, the more the model is learning when we have many root variations. We take a different 50, 210 images and train with the existing model with 30 epochs in this phase. Figures 9 and 10 visualize the accuracy and loss of the model in phase 2 respectively.
At the beginning of the training stage, we observe that train root accuracy drops from 85% to 80%. Not only train root accuracy, but all other categories' accuracy also drops after adding a new subset of images. The opposite behavior is observed in terms of the loss function. In epoch 0, loss functions of every category are increased. However, the final result of training 2 ends up with better train root, vowel, and constant accuracy of 92%, 95%, and 96%, respectively. In terms of the loss function, we observe the decrements over epochs in every case. These results imply that the model is appropriately converged and trained well due to more subset images used in the training process.
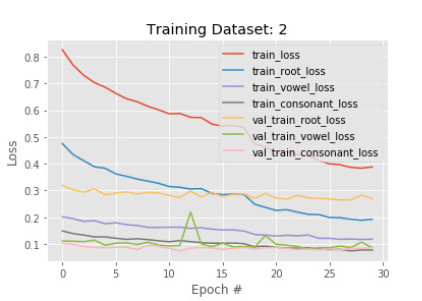
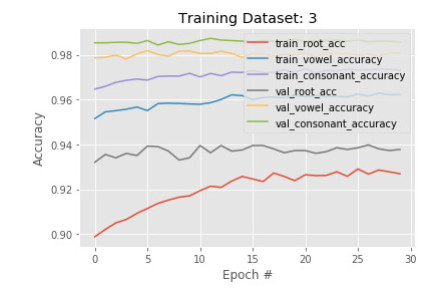
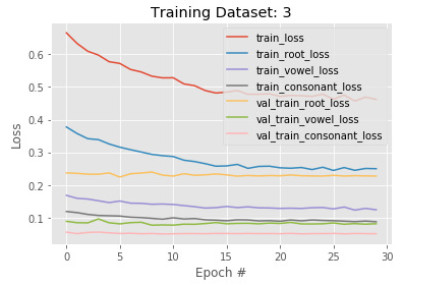
6.3. Phase 3 results
As a good result is maintained in our previous phases, we introduce another set of images with more variations in learning by our model. However, this time we observe little changes happen after the training. Figures 11 and 12 show the accuracy and loss of the model in phase 3. After another 30 epochs, training root, vowel, and constant accuracy are 94%, 96%, and 97%. The model root accuracy is increased by 2% and vowel, and constant accuracy are increased by 1%. The same behavior is found on the validation data also. It implies the model has converged very well and can be finalized by another training.
6.4. Final phase results
This final phase verifies the converge of accuracy and loss function by just doing another final training. Another set of 50, 210 images of Bangla handwritten letters are introduced. Figures 13 and 14 show the accuracy and loss function of the model in the final phase. The results show that root accuracy drops from 94% to 93% in the training stage, and the accuracy drops from 98% to 97% in the validation stage. Nevertheless, in all other cases, it seems improvement or no change. Also, the results start to create bumpy behavior in the accuracy metric, and loss functions are also converged. All the validation loss functions are 3% or below. These all results imply, our final model is converged and ready to report the final accuracy and loss.
7.
Conclusions
Despite the advances in the classification of grapheme images in computer vision, Bengali grapheme classification has mainly remained unsolved due to confusing characters and many variations. Moreover, Bangla is one of the most spoken languages in Asia, and its grapheme classification has not been made, as there is no application as yet. However, many industries like banks, post offices, book publishers, and many more industries need the Bangla handwritten letters recognition.
In this paper, we implement a CNN architecture with encoder-decoder, classifying 168 grapheme roots, 11 vowel diacritics (or accents), and 7 consonant diacritics from handwritten Bangla letters. One of the challenges is to deal with 13, 000 grapheme variations, which are way more than English or Arabic grapheme variations. The performance results show that the proposed model achieves root accuracy of 93%, vowel accuracy of 96%, and consonant accuracy of 97%, which are significantly better in Bangla grapheme classification than in previous research. Finally, we report the detailed loss and accuracy in 4 phases of training and validation to show how our proposed model learns over time.
To illustrate the model performance, we compared our model with a traditional CNN applied to the same dataset. The results show that the accuracy and loss function fluctuate over time in the traditional CNN model, which means an over-fitted model. In comparison, we see that the proposed CNN model with encoder-decoder does much better in classifying Bangla handwritten grapheme images.
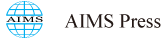








 DownLoad:
DownLoad: